Research Article
Isolation and Characterization of Extracellular L-Asparaginase Producing Bacillus Species from Morinda citrifolia Phyllosphere
Meghana NK, Navya MN, Harsha K and R. Aswati Nair*
Department of Biochemistry and Molecular Biology, Central University of Kerala (CUK), India
*Corresponding author: Nair AR, Department of Biochemistry and Molecular Biology, Central University of Kerala (CUK), Kasaragod, Kerala- 671 320, India, Telephone: +91 467 2309343, Fax Number: +91 467 2232402; E-mail: aswati@cukerala.ac.in
Copyright: © Meghana NK, et al. 2020. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Article Information: Submission: 05/11/2020; Accepted: 11/12/2020; Published: 14/12/2020
Abstract
Bacterial isolates were identified from phyllosphere of the medicinal plant, Morinda citrifolia, a source for the anti-neoplastic L-Asparaginase used as
a chemotherapy drug in lymphoblastic leukaemia. Amongst the isolates with significant anti-oxidant activities, seven were identified as L-Asparaginase
producers. Quantitative estimation of L-Asparaginase activity by the seven phyllospheric isolates revealed maximal specific activity for isolates designated,
McTTL3 (2.98 U. μg-1 protein) and McTUF8 (2.94 U.μg-1 protein). Molecular characterization of the selected phyllospheric isolates using 16S rDNA identified
isolate McTUF8 to Bacillus subtilis strain BcX1 (97.19% identity) and McTTL3 to Bacillus amyloliquefaciens subsp. plantarum strain Hk3-1 X030 (99.26%
identity). Further precipitation of L-Asparaginase from McTTL3 using [(NH4)2SO4] (20- 80% w/v) yielded 100-fold increase in specific activity (331.55 U.μg-
1 protein) at 20% saturation. The phyllospheric isolate designated McTTL3 identified in present study thus constitutes a potent source for commercial
production of the anti-neoplastic L-Asparaginase.
Keywords
Morinda citrifolia; L-Asparaginase; Phyllosphere; Bacillus
Introduction
L-Asparaginase (E.C. 3.5.1.1) is an enzyme of high therapeutic value
due to its potency as a chemotherapeutic drug in treatment of acute
lymphocytic leukemia [1-3]. The enzyme is present in many animal
tissues, in the serum of certain rodents, bacteria and plants except
humans [4]. L-Asparaginase catalyzes the deamination of asparagine
into aspartic acid and ammonia. A reduction in L-asparagine, which
tumour cells are unable to synthesise, accounts for its clinical action
by depriving tumor cells of L-asparagine. L-Asparaginases currently
in clinical use for chemotherapy include Elspar sourced from
Escherichia coli and Erwinaze from Erwinia chrysanthemi [5]. There
is an ongoing search for new L-Asparaginases with more desirable
properties due to rapid clearance of the enzyme from blood stream
and its L-glutaminase-dependent neurotoxicity [6]. Many potential
L-Asparaginase producing microbes have been identified and include
Erwinia cartovora [7], Enterobacter aerogenes [8], Corynebacterium glutamicum [9], Candida utilis [10], Staphylococcus aureus and Thermus thermophilus [11,12]. Bacterial source have been recognized
as most efficient as they are amenable to submerged fermentation in
batch and fed-batch bioreactors [5]. Much research interest has been
elicited towards identifying microbial isolates capable of producing
extracellular L-Asparaginase that would be safer, cost effective and
serologically different from current available L-Asparaginases [13].
Microbes colonizing plants especially those with medicinal
properties and anti-cancer potential have been identified to produce
L-Asparaginase [14,15]. In plants, microbes exist as epiphytes on
phyllosphere or above ground plant parts and as endophytes inside
the plant tissues [16-18]. In-depth sequencing of phyllosphere
microbes has revealed the microbiota, to be plant species-specific
[19,20]. Phyllosphere microbes play an important role in plant growth
and defense [21]. They have been identified as a potential source for a
number of metabolites and secondary metabolites such as antibiotics,
antitumor compounds and plant growth inducing factors [22,23].
Morinda citrifolia belonging to family Rubiaceae, also known
as Indian mulberry is a medicinal plant used in folk remedies by
Polynesians for over 2000 years [24]. The plant exhibits a broad range
of therapeutic effects that include anti-microbial, analgesic, hypotensive,
anti-hyperglycemic, nephron-protective, anti-inflammatory
and anti-tumor [25-27]. Fruit juice of M. citrifolia commercialized as
a potential nutraceutical with anti-tumor activity has been suggested
as a supplemental agent in cancer treatment along with sub-optimal
dose of standard chemotherapeutic agents like Adriamycin, cisplatin,
5-fluorouracil and vincristine [28,29]. Though earlier studies had
isolated fungal endophytes with anti-cancer activity from M. citrifolia
[28,30], similar characterization of phyllopsheric microbes is yet to be
undertaken. Present study characterized the phyllospheric microbes
of M. citrifolia and aimed towards (i) determination of anti-oxidant
activity; (ii) screening for L-Asparaginase production and (iii)
identification of L-Asparaginase producing microbes by 16S rDNA
sequencing.
Materials and Methods
Microbial cultures:
Pure cultures of ten phyllospheric bacteria (McTRF4; McTRF5;
McTRF7; McUTRF5, McTUF7, McTUF8, McUTUF4, McTTL3,
McTTL7 and McUTTL4) isolated in earlier studies from M. citrifolia
[31] and maintained on 20% (v/v) glycerol were sub-cultured in
Tryptone Yeast Extract (TYE) medium [20 g MgSO4, 0.2g CaCl2, 5 g
Tryptone, 3 g yeast extract and 3% NaCl (pH 7.0)] [31]. Preliminary
screening of the isolates for anti-oxidant activity was carried out by
plating the isolates on Tryptone yeast extract agar medium [20 g
MgSO4, 0.2 g CaCl2, 5 g Tryptone, 3 g yeast extract, 3% (w/v) NaCl,
pH 7.0] and incubating at 37 oC for 24 hours. Whatman No.1 filter
paper was placed over the plates for 24 hours to transfer the colonies.
To the filter paper, DPPH (1,1-diphenyl 1-2-picryl hydrazyl) solution
in ethanol (80 μg/ml) was sprayed and allowed to dry at an ambient
temperature.DPPH free radical scavenging activity:
Free radical scavenging ability of extracts from these isolates were determined by ability of extracts to bleach DPPH (1,1- diphenyl 1-2-picryl hydrazyl). For the same, isolates cultured overnight in TYE medium were centrifuged to pellet the cells. Supernatant was extracted thrice with ethyl acetate, concentrated to dryness and
dissolved in methanol (1 mg/ml). To 1.5 ml extract, equal volume 0.1 mM DPPH was added and incubated for 30 minutes at room temperature. DPPH decolourisation was measured at 517 nm with L-ascorbic acid as standard. Percentage DPPH scavenging activity was calculated as: DPPH scavenging activity (%)= [(Acontrol- Atest) x Acontrol-1] x 100, with Acontrol and Atest being absorbance of control and
test samples respectively.Screening isolates for L-Asparaginase production:
Primary screening of phyllospheric microbes for L-Asparaginase
production was carried out by rapid plate assay in M9 minimal
medium containing L-Asparagine as sole nitrogen source and using
phenol red as pH indicator which is yellow at acidic pH and pink
under alkaline conditions [32]. Briefly cultures were grown on M9
minimal media (2 g KH2PO4, 1 g MgSO4. 7H2O, 1 g CaCl2.2H2O, 3 g glucose, 20 g agar; pH 6.2) supplemented with 6 g L-Asparagine and
containing phenol red (2.5% v/v). Cultures were incubated overnight at 37 oC and positive isolates were inoculated on M9 minimal plate for secondary screening. Diameter of pink color zone was measured
at regular intervals ranging from 1-48 hours of incubation. Enzyme index was calculated according to the equation: Enzyme index = Pink zone (mm)/Colony diameter (mm).Determination of L-Asparaginase activity:
Isolates positive for L-Asparaginase production in log phase of growth were inoculated in M9 broth and incubated overnight at 180 rpm at 37 oC. Cell free extract was obtained after centrifugation of cultures at 10000 rpm for 10 minutes at 4 oC. L-Asparaginase activity was quantified in the supernatant by determining ammonia formation using Nessler’s method [33]. Briefly the reaction mixture (2 ml) containing 0.1 ml enzyme, 0.5 M Tris HCl buffer (pH 8.5) and 0.04 M L-Asparagine was incubated for 10 min at 37 oC. Reaction was terminated by adding 0.5 mL of 1.5 M Tri Chloro Acetic acid (TCA) and the mixture was then centrifuged at 10000 rpm for 10 min. To the supernatant, added 1 ml 1 N NaOH and 0.2 ml 0.1 M EDTA, incubated for 2 minutes, added 0.2 mL Nessler’s reagent and absorbance measured using a spectrophotometer at 450 nm after 10 minute incubation. Protein content was estimated using bovine serum albumin as standard (20-100 μg/ml). Activity was determined using ammonium sulphate reference standard (1-13 mM) [34]. One unit of L-Asparaginase (IU) activity was calculated as μmoles of ammonia released per 10 minutes and μg protein in reaction conditions at 37 ºC and pH 7.4.The M9 fermentation broth of positive isolate was centrifuged at
10,000 rpm for 30 minutes to remove the biomass. L-Asparaginase
was precipitated using ammonium sulphate [(NH4)2SO4] at different
saturation (10-80%). Precipitates were collected by centrifugation
at 10,000 rpm for 20 minutes at 4 °C. The obtained precipitate was
resuspended in a minimal volume of 1 M Tris HCl (pH 7.5).
Genotypic characterization of L-Asparaginase producers: Genomic DNA was isolated from the four isolates identified
positive for L-Asparaginase production. The extracted
DNA from each bacterial isolate was used as a template for
amplification of the 16S rRNA gene using the universal primers,
16s for (5′-CCAGCAGCCGCGGTAATACG- 3′) and 16sRev
(5′-ATCGG(C/T) TACCTTGTTACGACTTC- 3′). The obtained
sequences for the selected isolates were aligned and subject to
homology searches with BLAST algorithm at NCBI database (http://
blast.ncbi.nlm.nih.gov/Blast.cgi).
Results and Discussion
Phyllosphere is colonized by numerous microbes with the
composition of epiphytic microbes varying based on plant species
and environment [35]. Present study identified phyllospheric
L-Asparaginase producing isolates from Morinda citrifolia,
medicinal plant known for its anti-cancer activity. Medicinal plants
with anti-cancer activities are considered ideal source for isolating
microbes producing L-Asparaginase [36]. Phyllospheric microbes
display defensive effect on host plants by emitting volatile organic
compounds (VOCs) [37]. The volatiles can trigger production of
anti-oxidant metabolites [38]. Preliminary qualitative screening
revealed formation of white zones around the colonies against purple
background confirming selected isolates to exhibit antioxidant
activity. Two isolates viz., McTRF4 and McTTL3 from phyllosphere
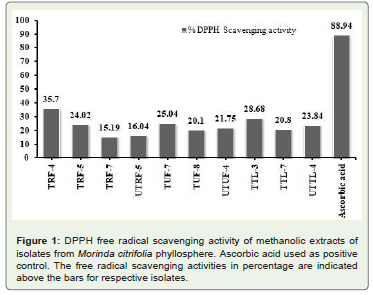
of M. citrifolia showed considerable anti-oxidant activity (Figure 1).
Isolates with significant anti-oxidant activities will be potential
producers of L-Asparaginase [39,40]. Preliminary screening in
M9 media revealed seven out of the 10 isolates as L-Asparaginase
producers. Further secondary screening by plate assay detected
formation of pink coloured zone around the colonies due to
change in pH to alkaline caused by release of ammonia during
deamination of L-Asparagine by L-Asparaginase. Measurement
of pink zones at regular time intervals (4-48 hrs) identified
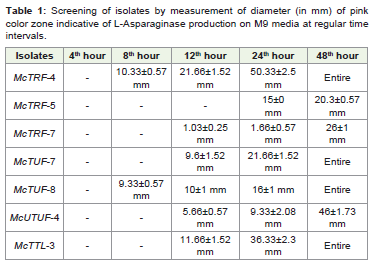
isolates, McTRF4, McTUF7, McTUF8 and McTTL3 to yield
maximum zone of activity within 48 hours of incubation (Table 1).
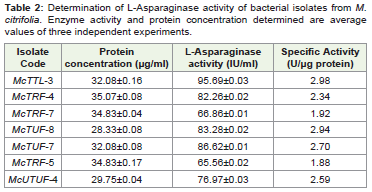
Quantitative estimation of L-Asparaginase activity by the seven
phyllospheric isolates revealed maximal specific activity for isolates,
McTTL3 (2.98 U. μg-1 protein) and McTUF8 (2.94 U. μg-1 protein)
(Table 2). Molecular characterization of isolates, McTTL3 and
McTUF8 displaying anti-oxidant activity and yielding maximal
L-Asparaginase production using 16S rDNA yielded a product of
molecular size 1500 bp. Sequencing and homology searches using
BLAST algorithm identified isolate, McTUF8 to Bacillus subtilis strain
BcX1 (GenBank Accession number: JX504009.1; 97.19% identity) and
McTTL3 to Bacillus amyloliquefaciens subsp. plantarum strain Hk3-1 X030 (GenBank Accession number: JF899255.1; 99.26% identity).
Bacillus species are ubiquitous in various ecological niches and are
known to produce various bioactive metabolites with a broad spectrum
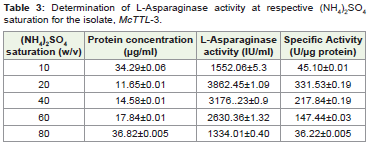
of activities [41]. Precipitation of L-Asparaginase using [(NH4)2SO4]
(20-80% w/v) from isolate, McTTL3 yielding maximal production
yielded high L-Asparaginase activity (331.53 U.μg-1 protein) at 20%
saturation (Table 3). A 100- fold increase in specific activity was
thus observed following [(NH42SO4] precipitation (Table 3). The
experiments revealed higher L-Asparaginase production by McTTL3 compared to earlier reports from bacterial and fungal endophytes.
For example, endophytic Fusarium spp. yielded 0.08-3.14 U. mL-1
[42], while an endophytic Penicillium spp. yielded 3.75 U. mL-1 [43].
Furthermore, isolates producing extracellular L-Asparaginase are
preferred over intracellular ones owing to enhanced production in
culture medium and ease of purification [44,45]. Thus the isolate
McTTL3 is a promising source for L-Asparaginase with applications
in food and pharmaceutical industry.
igure 1: DPPH free radical scavenging activity of methanolic extracts of
isolates from Morinda citrifolia phyllosphere. Ascorbic acid used as positive
control. The free radical scavenging activities in percentage are indicated
above the bars for respective isolates.
Table 1: Screening of isolates by measurement of diameter (in mm) of pink
color zone indicative of L-Asparaginase production on M9 media at regular time
intervals.
Table 2: Determination of L-Asparaginase activity of bacterial isolates from M.
citrifolia. Enzyme activity and protein concentration determined are average
values of three independent experiments.
Conclusion
Present phyllospheric isolate designated McTTL3 identified in
present study as B. amyloliquefaciens constitutes a potent source
for large scale production of the anti-neoplastic L-Asparaginase.
Endophytic B. amyloliquefaciens producing L-Asparaginase have
been identified from medicinal plants like Ophiopogon japonicas and
Curcuma amada [46,47].
Acknowledgements
Authors acknowledge the research facilities at CUK for
undertaking the present work. HK acknowledges UGC for the Junior
Research Fellowship (JRF).