Research Article
Morphological and Molecular Characterization of Banana in Kerala, India
Anu A1, Geethalakshmi S1* and Vazhackarickal PJ2
1Department of Biotechnology, Sree Narayana Guru College, India
2Department of Biotechnology, Mar Augusthinose College, India
*Corresponding author: Geethalakshmi S, Department of Biotechnology, Sree Narayana Guru College, Coimbatore,
641105 Tamil Nadu, India; E-mail: s.geethalakshmi@gmail.com
Copyright: © Anu A, et al. 2019. This is an open access article distributed under the Creative Commons Attribution License, which
permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Article Information: Submission: 23/10/2019; Accepted: 25/11/2019; Published: 27/11/2019
Abstract
Banana is one of the most important food crops all over the world. There are around 365 varieties of bananas available throughout the world. Banana
is a traditional medicine for diabetes, cancer, diarrhea and also highly nutritional food crop. In this study, commonly used varieties of banana are taken for
characterization by morphology and genotype which is based on International Plant Genetic Resources Institute, 1984 (Descriptors for banana (Musa sp.,)
and RAPD analysis. Five varieties were morphologically similar in parameters such as leaf habit, pseudo stem appearance and peel color. RAPD analysis
proved that these varieties of banana are closely related which coincides with the morphological characterization.
Keywords
RAPD analysis; Morphological characters; Genotype
Introduction
Banana is naturally packed with nutrients, fibers, protein
and other compounds. Various parts of plantain tree are used for
medicinal purpose and as a food product. Banana is a monocotyledon
plant in the genus Musa (Musaceae). They are giant herbs, commonly
up to 3m in height and their stem is not as strong as other trees.
Morphologically, each variety is different from others in parameters
such as leaf habit, male bud size, color pedicle size, pedicle position,
fruit numbers, pulp color, mature peel color and immature peel color.
Due to this variation, Musa has been taxonomically classified to 50
species. Most of the edible bananas are from derived from two wild
species like M. acuminata (having AA genome) and M. balbisian
(having BB genome) through inter and intra specific hybridization,
resulting in the generation of many genome groups such as
AA,AB,AAA,AAB,ABB,AABB,AAAB and ABBB [1]. It is considered
least because of the widespread vegetative reproduction and natural
occurrence of many hybrids varieties [2]. Climate and soil fertility
influences the growth of banana.
Differentiating varieties of banana based on their morphology has
some limitations in the accurate identification because limited traits
are available for characterization [3]. Molecular markers are used for
studies such as AFLP, RFLP, RAPD, ISSR, SSR, and SNP for accurate
analysis. Random Amplified Polymorphic DNA (RAPD) technique is
widely used for population study and genetic linkage because of their
high polymorphic nature [4], frequent occurrence in the genome, easy
access, fast process and easy exchange of data between laboratories.
RAPD molecular markers are widely used for variety of species
identification for apple (Malus species L), grapes (Vitis) and Rice
(Oryza sativa L). It is an efficient and inexpensive technique without
requiring prior knowledge of the genome [5]. RAPD analysis depends
on the selection of primers; when five different primers are used for
the selection process, it is helpful to identify which primers contain
high polymorphic activity which is provides useful amplification
products.
Materials and Methods
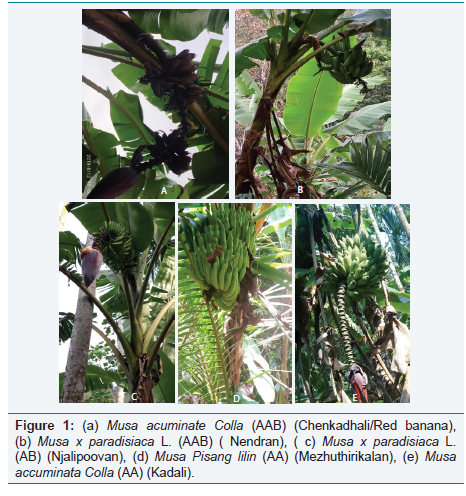
Five banana fruits samples and leaf samples were collected from Palakkad. They were identified as S1: Musa acuminate Colla (AAA) (Chenkadhali/Red banana), S2: Musa x paradisiacal L. (AB) (Njalipoovan), S3: Musa Pisang lilin (AA) (Mezhuthirikalan), S4: Musa accuminata Colla (AA) (Kadali) and S5: Musa x paradisiaca L. (AAB) (Nendran). The samples were authenticated by Botanical Survey of India and Banana Research Centre, Kannara, Kerala (Figure 1).
Morphological characterization:
The collected five varieties of banana were morphologically
analyzed for their features based on the IPGRI 1984, descriptors of
banana. Leaf habit, leaf colour, pseudo stem colour, size, pedicle size,
color, appearance, male bud color, shape, fruit color, shape, size were
observed and tabulated.Molecular characterization of Banana:
RAPD is a PCR based method to detect variation between
individuals of species by selective amplification of some polymorphic
sequences in their genomes. RAPDs are of much use to construct
genetic map [6].Isolation of DNA from banana:
DNA was isolated from the leaf samples using NucleoSpin ®
Plant II kit ((Macherey-Nagel) as per manufacturer’s instruction. The
eluted DNA was stored at 4 °C.Agarose gel electrophoresis:
Agarose gel electrophoresis was performed according to standard
protocol (Molecular Cloning -CSHL Press) to check quality of the
isolated DNA. The samples were loaded to 0.8% agarose gel prepared
in 0.5X TBE buffer containing 0.5 μg/ml EtBr. After electrophoresis
the gel was viewed on a gel docking station and photographed.
Figure 1: (a) Musa acuminate Colla (AAB) (Chenkadhali/Red banana), (b) Musa x paradisiaca L. (AAB) ( Nendran), ( c) Musa x paradisiaca L. (AB) (Njalipoovan), (d) Musa Pisang lilin (AA) (Mezhuthirikalan), (e) Musa accuminata Colla (AA) (Kadali).
RAPD analysis:
Restriction enzymes and primers were purchased from Medox
Biotech, India. PCR amplification was performed in two stages. The
pre-selective amplification was performed with an amplification
profile of 95 °C for 5 minute, annealing at 42 °C for 1min, extension
at 72 °C for 1.3 min, repeated for 34 cycles, then extend at 72 °C for
10 min. Further amplification was performed with a cycling profile
of 94 °C for 0.45 min, 42 °C for 1.00 min, 72 °C for 1.30 min cycles
followed by extension at 72 °C for 10 min and a cooling of 4 °C for 30
min. Electrophoresis of the PCR products was carried out on agarose
gel (1.2%), by loading 10 μl of each DNA samples .The electrophoresis
was run at 75 V for 3 hrs and viewed on a gel docking station and
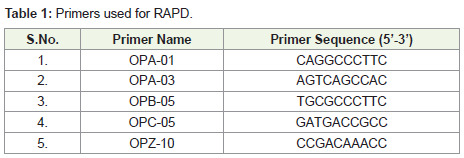
photographed (Table 1).Data scoring:
The standard of scoring was generated for most dominant DNA
markers and only clear distinctive bands were scored using NTSYS
software. Bands were recorded into binary symbols “1” for band
presence and “0” for band absence using and phylogenetic tree was
constructed using NTSYS software.Results and Discussion
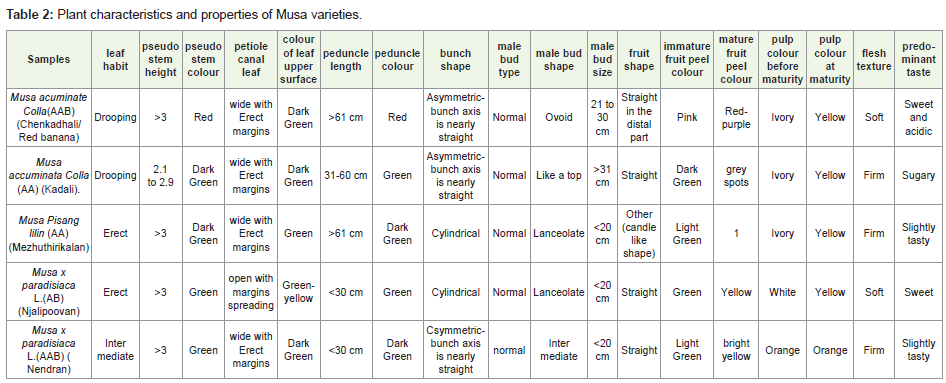
The five banana samples collected from palakkad district were morphologically characterized based on several parameters are tabulated in (Table 2). From this table, it can be seen that the morphological parameters of all the samples are similar up to 75%. This is confirmed by the phylogenetic tree which forms the first out group of Musa x paradisiaca L.(AB) (Njalipoovan) and Musa x paradisiaca L.(AAB) (Nendran) second out group of (Chenkadhali/Red banana) Musa accuminata Colla (AA) (Kadali/Kali). Musa Pisang lilin (AA) (Mezhuthirikalan/ sudhari) is found common between these two groups [7] (Figure 2).
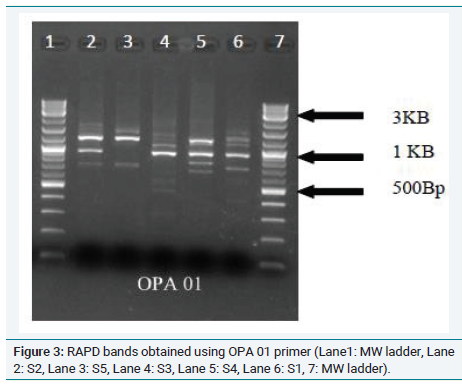
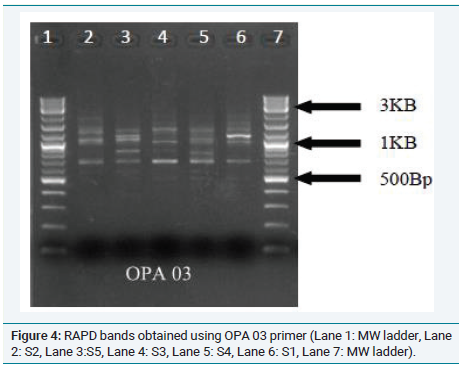
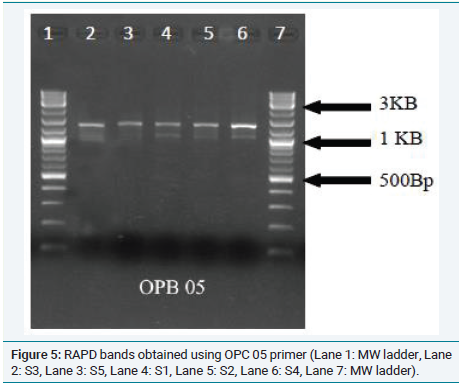
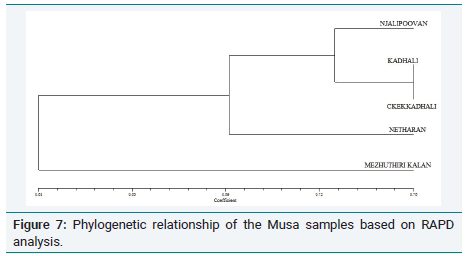
In order to further confirm the relationship between the samples, RAPD analysis was performed and the phylogenetic tree is shown in (Figure 3). This tree confirms the grouping of Musa acuminate Colla(AAB) (Chenkadhali/Red banana) Musa accuminata Colla(AA) (Kadali) and a close relationship between Musa x paradisiaca L.(AB) (Njalipoovan) and Musa x paradisiaca L.(AAB)( Nendran) (Figure 4 and 5).
http://www.opensciencepublications.com/fulltextarticles/images/JPSR-2349-2805-6-188-fig2.png:
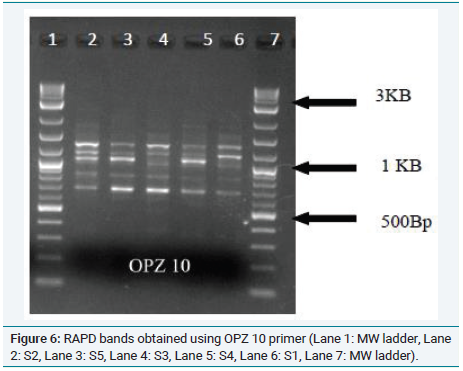
Figure 2: Phylogenetic relationship of morphological parameters of Musa samples.From these results, it can be concluded that genotypic relationship between the samples also explicitly exhibited in their phenotype. It is well known that Musa paradisiaca L.(Nendran) is effective in curing breast cancer [8]. Since Musa x paradisiaca L.(AB) (Njalipoovan) shows close relation with Musa x paradisiaca L.(AAB)( Nendran). Both morphologically and genotypically it is expected that this variety also has an anticancer potential (Figure 6 and 7). Further studies are needed to confirm the gene expression pattern in the collected varieties which can be used for pharmaceutical application [9].
Figure 3: RAPD bands obtained using OPA 01 primer (Lane1: MW ladder, Lane
2: S2, Lane 3: S5, Lane 4: S3, Lane 5: S4, Lane 6: S1, 7: MW ladder).
Figure 4: RAPD bands obtained using OPA 03 primer (Lane 1: MW ladder, Lane
2: S2, Lane 3:S5, Lane 4: S3, Lane 5: S4, Lane 6: S1, Lane 7: MW ladder).
Figure 5: RAPD bands obtained using OPC 05 primer (Lane 1: MW ladder, Lane
2: S3, Lane 3: S5, Lane 4: S1, Lane 5: S2, Lane 6: S4, Lane 7: MW ladder).
Figure 6: RAPD bands obtained using OPZ 10 primer (Lane 1: MW ladder, Lane
2: S2, Lane 3: S5, Lane 4: S3, Lane 5: S4, Lane 6: S1, Lane 7: MW ladder).
Acknowledgement
The authors are grateful to the Staff of Department of
Biotechnology, Sree Narayana Guru College for necessary support.