Research Article
Investigating the Antiphage Activity of the Methanolic Extract of Aegle marmelos against Lactic Acid Bacteriophages
Mulay R and Bhathena Z*
Department of Microbiology, Bhavan’s College, Andheri West, Mumbai, India
*Corresponding author:Zarine P Bhathena, Department of Microbiology, Bhavan’s College, Andheri West, Mumbai, Maharashtra, India.Email Id: zarine_bhathena@rediffmail.com
Copyright: ©Mulay R, et al. 2025. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Article Information:Submission: 02/01/2025; Accepted: 29/01/2025; Published: 04/02/2025
Abstract
Bacteriophages are common nuisance viruses that cause tremendous loss to the dairy industry by lysing lactic acid bacteria. Thus, an intervention using inclusion of ayurvedic herbs to prevent such lysis of potential lactic acid bacterial strains was set up. In this study, a thorough phytochemical analysis
of methanolic extract of Aegle marmelos leaf powder undertaken showed the presence of major groups of secondary metabolitesin the extract while LCMS and GCMS identified various bioactive compounds in it. Antiphage assay using crude methanolic extractdemonstrated a significant reduction in P001 phage with a log reduction of 0.0846 ± 0.083 after 20 min of exposure and 0.708 ± 0.081 after 90 min of exposure for the extract without inhibiting the probiotic host culture. These findings offer valuable insights into the extract’s potential for protecting the viability of the probiotic host culture in dairy applications.
Keywords:Antiphage Activity; Bael; Lactococcus lactis; Dairy Bacteriophages
Introduction
Aegle marmelos is a plant native to India belonging to the family
Rutaceae. It is an important medicinal plant used in traditional Indian
Ayurvedic medicine. It has various names in colloquial language such
as Bengal quince, Indian quince, Holy fruit, Golden apple in English,
“Bilva” in Sanskrit while in Hindi it is known as “Bael”. According
to Charaka (1500 BC), Bael is considered as an emblem of fertility
and is a holy tree capable of healing and strengthening the body.
Diverse ethnomedical properties of Bael have been reported in the
pharmacological studies which include antimicrobial, antifungal,
anti-inflammatory, antipyretic, hypoglycemic, antidyslipidemic,
immunomodulatory, antiproliferative, wound-healing, insecticidal,
anticancer, antidiabetic, and cardioprotective properties[1].This
plant contains a diverse groups of bioactive compounds like alkaloids,
saponins, tannins, cardiac glycosides, flavonoids, and phytosterols
which provide medicinal properties to the plant [2]. The major
phytochemicals reported in Bael include Skimmianine, Aegeline,
Lupeol, Cineol, Citral, Citronella, Cuminaldehyde, Eugenol,
Marmesinine, Marmelosin, Luvangetin, Aurapten, Psoralen, and
Marmelide[3]. All the parts of the Aegle marmelos plant including
fruits, stem, bark, and leaves exhibit medicinal properties but the most
health benefits associated with Bael are attributed to the numerous
phytocompounds present in its leaves [4].
Fermented foods have received attention worldwide for their
varied health benefits. Lactic acid bacteria are widely used in food
industry, particularly in dairy industry to manufacture a range
of fermented foods [5]. However, their use in dairy industry is
encountered with several challenges, most importantly being
attacked by bacteriophages which lyse them causing fermentation
disturbances [6]. Lytic phages are known to infect probiotic starter
cultures in commercial manufacturing facilities leading to loss
of productivity. Moreover, they are resistant to pasteurization
temperatures and traditional phage control measures [7]. Thus, by
conducting in-depth studies on these phages, targeted strategies to
prevent their infections in commercial manufacturing plants can be
devised. This study thus plans to evaluate the potential of methanolic
extract of Aegle marmelos against Lactococcus P001 phage along with
its thorough phytochemical profiling.
Materials and Methods
Procurement of Plant material:
Complete plant of Aegle marmelos was procured from the
Botanical Garden & Nursery within Bhavan’s College Campus,
Mumbai and confirmed of its authenticity by Botanists at Department
of Botany, Bhavan’s College. The leaves of the plant were dried
and ground into fine powder using a grinder and stored in airtight
containers until further use.Preparation of plant extract and Phytochemical analysis:
5 grams of the powdered material was dissolved in 50 ml methanol
[Loba Chemie] and kept on a rotary shaker for 3 days. Every day, the
shaker was switched on for 8 hours thus ensuring 24 hours of effective
shaker treatment. Each extract was then filtered by whatmann paper
No 1 [Himedia] to separate the powder. Preliminary qualitative
testing for alkaloids, phenolic compounds, tannins, flavonoids,
saponins along with quantitative testing for phenolic compounds,
alkaloids and flavonoids,was conducted using standard protocols [8-11].HPTLC fingerprinting of the extract:
HPTLC fingerprinting of the extract was carried out using
mobile phase consisting of ethyl acetate: water: formic acid:
acetic acid (100:26:11:11- v/v/v/v). The stationary phase
consisted of TLC silica gel 60 F254 (Merck -100 X 100 mm).
Specified volumes of sample was applied on to the TLC
plates using Linomat 5 (CAMAG), semiautomatic sample
dispenser. The TLC plates were scanned by CAMAG Scanner
4 which had a 6 X 0.45 mm micro-slit dimension and a scan
speed of 20mm/sec and data resolution of 100μm/step. The
developed chromatogram was visualised at 254 nm, 366
nm, and white light after derivatization with anisaldehyde
sulphuric acid reagent and Natural Product A reagent and
photodocumented.Liquid Chromatography Mass Spectrometry (LC-MS) Analysis:
LCMS analysis of the extract was carried out using Q-TOF
mass spectrophotometer with Dual AJS ESI (G6550A, Agilent
Technologies, USA). The mobile phase consisted of 0.1 %
formic acid (A) and methanol (B). The analysis followed a
linear gradient program where in initial conditions were
solvent A 95%:B 5%; 0-25 min, changed to solvent A 0%:B
100%; 25-30 min and back to solvent A 95%:B 5%; 31-35 min.
The flow rate was maintained at 0.3 ml/min and injection
volume was 5 μL and the column used was Infinity HPLC
G1316C (Agilent, USA). The data generated was processed,
analysed, and interpreted using Mass Hunter software
(Agilent, USA).Gas chromatography Mass Spectrometry (GC-MS) Analysis:
GCMS analysis of the extract was carried out on 7890B GC
system (Agilent, USA) connected to Jeol AccuTOF GCV with FID
detector and head space injector. (Agilent, USA). The stationary
phase column was HP-5 (30 mX0.32 mm, 0.25 μm) and helium was
used as a carrier gas at a constant flow rate of 1 mL/min. The oven
temperature was initially set at 60ºC for 1 min and then was increased
by 6ºC per min and maintained at 200ºC for 2 min. Final temperature
was maintained at 280ºC. The AccuTOF MS (Jeol) detector was used
for eluting the molecules enabling their detection. The total run time
was 28 min. The compounds eluted from the extract were identified
and characterized by employing standard spectral libraries like NIST.Preparation of plant extract for antiphage activity:
50 g of the powdered plant was dissolved in 300 mL methanol
[Loba Chemie, India] and kept on a rotary shaker for 3 days. Every
day, the shaker was switched on for 8 hr thus ensuring 24 hr of
effective shaker treatment. The extract was then filtered by whatmann
paper No 1 [Himedia, India] to separate the powder and the methanol
was allowed to evaporate. The residue was weighed and dissolved in
DMSO [Loba Chemie, India] and DMSO dissolved extract was used
for further experimentation.Procurement and maintenance of LAB host and its bacteriophage:
Lactococcus lactis [DSM-4366], a probiotic host culture used in the
cheese fermentation and its virulent bacteriophageLactococcus phage
P001 [DSM-4262] were obtained from DSMZ-German Collection
of Microorganisms and Cell Cultures, Germany. The cultures were
grown and maintained on M17 medium [Himedia, India] at 30ºC
under aerobic conditions.Determination of MIC against Lactococcus lactis host:
MIC of the extract against the Lactococcus lactis probiotic host
culture was performed by standard tube dilution technique to
quantify the absence of extract activity on the host strain. DMSO
dissolved extract was double diluted in M17 broth [Himedia, India]
in two sets. The first set was labelled as ‘test’ and the second set was
labelled as ‘control’. Post dilution, 50 μL of the actively growing host
was added to the ‘test’ set while 50 μL saline was added to the ‘control’
set. The tubes were incubated at 37ºC for 24 hr. Post incubation, 10
μL extract from all the tubes of ‘test’ and ‘control’ sets were spotted on
M17 agar plate (Himedia) and the plates were incubated at 37ºC for
24 hr. Post incubation, growth of culture at the spot was considered
as positive result. Appropriate controls were set up to eliminate false
results and each set was run in triplicates and repeated three times to
confirm the reproducibility of the result.Determination of antiphage activity [12]:
450 μL of the plant extract (at the dilution that did not inhibit the
probiotic host) was mixed with 50 μL of the Lactococcus phage P001
and incubated at 37ºC for different time intervals viz 20 min and 90
min. For control, 450 μL of St. saline was mixed with 50 μL of the
Lactococcus phage P001 and incubated at room temperature for same
time intervals. 100 μL of the test and control aliquots were then mixed
with 300 μL of actively growing Lactococcus lactis host culture and
subjected to plaque assay by standard double agar overlay method.
Post incubation, the plaques were counted the log reduction values
for two periods of exposure was calculated. Each set of exposure
times was run in triplicates and repeated three times to confirm the
reproducibility of the result.Statistical Analysis:
Statistical analysis was done using Prism Version 9.0 (Graph Pad
Software, Inc USA). Absorbance values of standards for estimation
of alkaloids, flavonoids, and phenolic compounds were entered into
the software yielding a calibration curve equation. This equation
was then used to interpolate absorbance values of the sample, and
the concentrations determined were expressed as mg/ml. The log
reduction values were presented as mean ± SD. To access the statistical
significance of the findings, a statistical level of P<0.05 was employed.Results and Discussion
The present study comprehensively investigates the phytochemical
composition of the methanolic extract of Aegle marmelos leaves and
its potential to prevent phage infection of Lactococcus lactis P001.
Qualitative phytochemical analysis confirmed the presence of key
secondary metabolites such as alkaloids, phenolic compounds,
reducing sugars, flavonoids and saponins. Tannins were not detected,
and the absence of tannins is particularly interesting as phenolic
compounds typically co-occur with tannins in many plant extracts. The
presence of these metabolites is significant as they have been reported
for anti-inflammatory, antiviral and antibacterial properties [13,14].
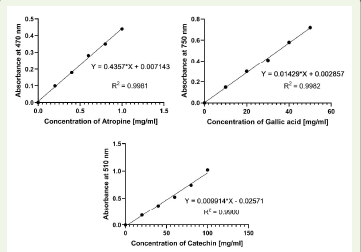
Alkaloid content was quantified using a standard curve plotted with
atropine, resulting in an equation Y=0.3457*X+0.02381(Figure 1). The
concentration of alkaloids was found to be 0.104 ± 0.04 mg of atropine/
mg. Phenolic content was determined using a calibration curve with
gallic acid, yielding the equation Y=0.01429*X+0.002857 [Figure 1].
The phenolic concentration was 19.75 ± 0.49 mg of gallic acid/mg.
Flavonoid content was quantified using catechin as a standard, with
a calibration curve equation Y=0.009914*X−0.02571[Figure 1]. The
Figure 1:Calibration curve for estimation of alkaloids using atropine
standard and phenolic compounds using gallic acid standard & flavonoids
using catechin standard.
flavonoid concentration was 54.03 ± 1.42 mg of catechin/mg. These
high levels of phenolic compounds highlight the strong antioxidant
potential of the extract [15].
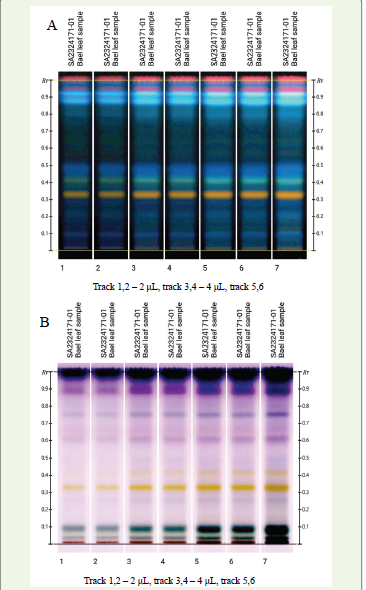
HPTLC fingerprint of the methanolic extract of Aegle marmelos
was developed using the mobile phase composed of ethyl acetate:
water: formic acid: acetic acid (100:26:11:11 v/v). After development,
the TLC plate was derivatized with universal derivatizing agents
anisaldehyde sulphuric acid and natural product A reagent revealing
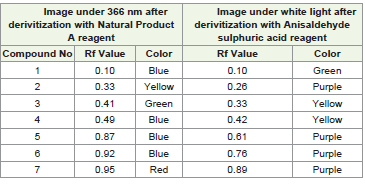
seven distinct bands. Each band corresponded to the separation of
a different compound with a unique Rf value as shown in [Table 1].
Specific identities of these compounds could not be definitively
determined without comparison to known standards. [Figure 2A]
represents the HPTLC fingerprint documented under 366nm light
following derivatization with natural product A reagent while [Figure 2B] shows the fingerprint under white light after treatment with
anisaldehyde sulphuric acid reagent.
Figure 2:A:Image under 366nm after derivatization with Natural Product A
reagent; B: Image under white light after derivatization with anisaldehyde
sulphuric acid
Advanced analytical techniques, specifically LC-MS and GC-MS,
were employed to gain deeper insights into the bioactive compounds
present in the extract. LC-MS analysis identified several significant
compounds, including alkaloids like piperine, which is known for
its potential antiviral properties against Dengue and Ebola viruses
as suggested by computational studies [16]. The presence of high
molecular weight compounds, such as triterpenoids and sterols, is
particularly relevant as these compounds are often associated with
enhanced bioactivity due to their structural complexity, which may
facilitate stronger interactions with biological targets, including viral
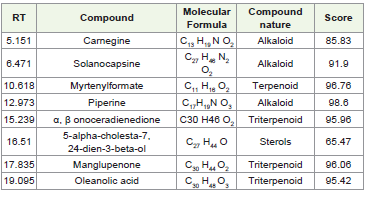
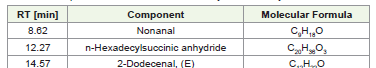
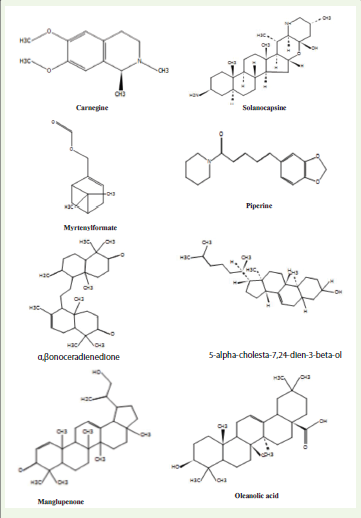
particles and phages. The detailed list of compounds identified by LCMS
is provided in [Table 3] , and the chromatogram obtained from
LC-MS analysis is shown in [Figure 3] and structures identified are
shown in [Figure 4].
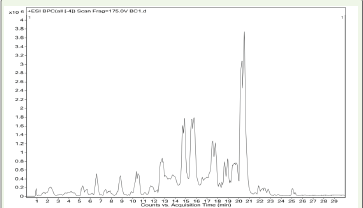
GCMS analysis revealed the presence of several compounds, each
identified based on its specific retention time, analysing their mass
spectra and corresponding m/z values using NIST spectral databases.
The chromatogram obtained from GC-MS analysis is shown in
[Figure 5], with the detailed list of identified compounds provided in
[Table 3] .
The determination of the MIC ensured that the plant extract did
not inhibit the probiotic host culture, Lactococcus lactis. The MIC
value was determined to be 80 mg/ml, with the highest concentration
tolerated by the host being 70 mg/ml. This concentration was then
used in subsequent experiments to evaluate the extract’s antiphage
activity, ensuring that the host culture remained viable and unaffected
by the extract.
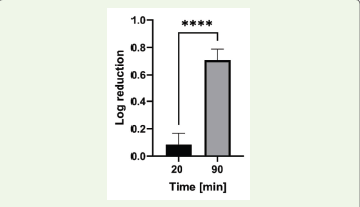
The antiphage activity was further evaluated by exposing the phage
Figure 6:Effect of methanolic extract of Aegle marmelos on Lactococcus
P001 phage (**** = p <0.0001).
to the extract at concentration of 70 mg/ml for two different durations,
20 minutes and 90 minutes. The results showed a significant timedependent
increase in antiphage activity, with a mean log reduction
of 0.0846 ± 0.083 observed after 20 minutes of exposure, which
increased to 0.708 ± 0.081 after 90 minutes of exposure [Figure 6].
The statistically significant difference in antiphage activity between
the two exposure times (p < 0.0001) suggests that the bioactive
compounds in the extract may interfere more effectively with
different stages of phage replication over extended exposure times.
This finding implies that prolonged contact with the extract enhances
its capacity to inhibit phage activity, potentially by disrupting key
processes in the phage life cycle.
These findings align with previous studies, including those
involving other plant extracts like Withania somnifera, where a log
reduction of 0.713 ± 0.08 after 20 minutes and 0.736 ± 0.18 after
90 minutes of exposure to the phage was observed, showing that
different plant extracts might share common mechanisms of action
against phages [17]. Other plant extracts, such as those from Phoenix
dactylifera [18] and Plantago major [19], have also been reported to exhibit significant antiphage activity, further supporting the potential
of plant-derived compounds in phage control strategies.
The current study provides valuable insights into the potential
applications of Aegle marmelos leaf extract in the dairy industry,
particularly in mitigating phage infections that can disrupt
fermentation processes. The extract’s ability to inhibit Lactococcus
P001 phage without adversely affecting the probiotic host highlights
its potential as a natural and effective solution to phage-related
challenges. This study opens avenues for the development of new,
medicinal plant-based strategies for phage control in industrial
applications.
Conclusion
Studying the phytochemical profile of Aegle marmelos leaves
extract highlighted the rich composition of varied secondary
metabolites including alkaloids, phenols, flavonoids, and saponins.
This study also demonstrated the effectiveness of the methanolic
extract for reducing the phage infection of P001 phage without
impacting the viability of the probiotic host culture suggesting its
application in preventing phage infection. Presently, our focus lies
on purifying the plant extract to separate the bioactive compounds,
elucidating their antiphage effects and understanding their precise
mechanisms.
Acknowledgements
The authors would like to extend their gratitude to the
Department of Microbiology, Bhavan’s College for providing the
necessary facilities and support for conducting this study. The authors
gratefully acknowledge Anchrome Labs Pvt Ltd for their support in
HPTLC fingerprinting and SAIF, IIT Bombay for their assistance in
LCMS and GCMS analyses.
Conflict of Interest:The authors declare no conflict of interest
in this study.