Review Article
A Contextual Analysis through Cellular Signaling Pathway, Immunogenicity and Mechanism of Action in T- Cell Activation on Gut Microbiome
Samanta D and Bhattacharya M*
Department of Biotechnology, Techno India University, EM- 4, Sector- V, Salt Lake, Kolkata- 700091, WB, India
*Corresponding author:Malavika Bhattacharya, Department of Biotechnology, Techno India University, EM- 4, Sector- V, Salt Lake, Kolkata, WB, India, Email Id:malavikab@gmail.com
Article Information:Submission: 01/07/2024; Accepted: 02/08/2024; Published: 05/08/2024
Copyright: ©2024 Samanta D, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
The gut, or digestive tract, is the main functional system of the human body that controls the intestine for accumulating gut microbes in the epithelial tissue. Intestinal immune cells are helping the gut microbiome environment to differentiate between intestinal epithelial barrier and immune function. The gut
microbiota, or microbial community of the intestine, is the active companion of human health and includes the necessary functions of the intestine along with the distal and proximal organs. The gut ecosystem depends upon bidirectional microbiota-host communication without any direct cellular contact. According
to the microbiota and host-derived extracellular vesicles (EVs), the main emphasis is given on performing inter-kingdom crosslinking. It is proven from previous accumulation of body that gut microbiota derived from bacterial secretion of certain vesicles helps to transport and deliver inside host cell effector
molecules, which causes the modulation of host cell signaling pathways and cellular programming. But the gut microbiota, which secretes vesicles, has efficient effects on healthy or diseased conditions of the body. The recent background on microbiota EVs for controlling host metabolism, intestinal barrier,
host pathogenic integrity, and immune training is highlighted in this study. [1]
The metabolic receptors of enigmatic inflammasomes in auto-immune diseases and crosstalk with innate immune regulators are dependent upon nucleotide binding domain and leucine rich repeat receptor (NLR). This NLR mediation of inflammatory activation is essential in host pathogenic response
and danger-associated molecular patterns (DAMPs)-related metabolic disease. Several cellular metabolic pathways can cause interaction with NLRs, and in contrast to negative regulation, tumorigenesis and autoimmune disorders interact with multiple innate immune receptors and disease modulation. In the host
pathogenic response, NLR activation is necessary in controlling metabolic pathways, which further target various levels of immune-metabolic diseases or syndrmes. The lesser known NLR studies of inflammasomes, which are activated by particular modes, further help to interact with metabolites and immune
receptors, but however, the function of the procession of metabolic diseases is not described thoroughly. So, this study is evidence of targeted NLR activity in metabolic pathways and crosslinking with immune receptor connections in GPCR signaling, gut microbiome, and also the complement pathways of the
immune system to understand the disease procedures. [2]
Keywords:Gut Microbiome; Bacterial Extracellular Vesicles (Bevs); Immunomodulation; GPCR Signaling; Homeostasis; Gastro-Intestinal Epithelium; Helicobacter Pylori
Introduction
The gut environment in the human body is largely colonized
by microbes that is all together called microbiome. In the intestinal
tract microbial colonies are excess in number i.e., 10 times
organisms per gram of wet mass of fecal composition in the colon.
The gut microbiota or symbiotic microbial community is generally
composed of symbiotic bacteria. In a combination gut microbiota
contains huge genes which conveys human genome encoded genetic
information. As a hidden organ gut microbiota contributes essential
host cell functions. Gut microbiota provides immunogenic capacity
of homeostasis and inflammation towards pathogenic interaction
along with nutritional, metabolic and energy mediated activities. The
gastrointestinal immune response has been associated with pivotal
functional activities of the gut microbiota. [3]
Mammalian host pathogenic interaction is involved with the gut
microbiota. This gut microbiome provides immunogenic protection
of synergistic recognition. Pathogen associated molecular patterns
are identified by pattern recognition receptors (PRRs) actively specify
innate immune cellular functions. But downstream signaling process
directly proceeds to phagocytic activities pathogens thus further
it regulates effector immune cells. The structural and functional
immune activity are in turn associated with the gut microbiome.
Antigen processing cells APCs and peptide epitopes help to engulf
foreign molecules and MHC class proteins are highly associated with
TCR affinity by high specificity. The peptide antigen involves with the
T cells activity for development of specificity and cellular or humoral
receptor activities. [4]
Many commensal bacteria are the source of enzymes neither
encoded by host nor intrinsic genome thus helping to the dietary
components in digestive system and accumulating supplementary
food items such as vitamins. Gut microbiota also protected from
pathogenic infection by intestinal pathogenic bacteria or gastroenteric
pathogens through nutrient competitive environment.
Mucus epithelium of intestine get immunity from gut microbiota.
Microbial colonies in lumen and epithelial tissues are subjected to
diversification by cellular vicinity. Mucosa in intestine is covered
with finger like bulging called villi and glandular septum called
crypts which accumulates nutrients and produce covered stem cell
background. The sub-epithelium of intestine is composed of stem
cells, goblet cells, neuroendocrine cells etc. [5]
Goblet cells secrets glycosylated mucins which further generates
mucus matrix in the surface of epithelium to form large or small bowel.
The luminal antigens are subjected to sub-epithelial APCs. The spongy
mucus layer generated by goblet cells is locally releasing antimicrobial
peptides in Paneth cells crypts. Apically situated microvilli exposed
vesicles by IECs that have been affecting antimicrobial function.
Catalytic activities of endometric vesicles secrets alkaline phosphatase
to detoxify harmful bacterial endotoxins and prevents clumping of
cells and also cease the cell division of entero pathogenic bacteria.
Absorptive cells of gastro-enteric epithelium are diversified from
gene expression and phenotypical characterization according with
proximal-distal axis of intestine along with their anatomical integrity
with different cell types or sub epithelial lymphoid structures, Peyer’s
patches, lymphoid follicles, FAE cells, M cells and phagocytes. [6]
The large and small intestine are structurally and functionally
different and separated between duodenum, jejunum, ileum,
caecum and colon. The composition is also varied from different
pH level, oxygen tension, retention time of luminal composition,
microbiota and bacterial capacity, density of immune stimulatory
microbial ligands, cellular density of mucus layer, regulating gastroenteric
antimicrobial peptides, anatomy and surface structure. Villi
structures, epithelial cell subtypes, enzyme secretion and metabolism
of trans epithelial cells and transport capacities with immune cell
subtypes. Intraepithelial lymphocytes (IELs) penetrate the epithelium
between phagocytosis and dendritic cells to the antigenic luminal
epithelial cells. [7]
Not only the microorganisms are present inside the human body
but also gastro enteric tract is contained with diversified number
of commensal, mutualistic or symbiotic bacteria mainly named
as bacteroidetes, actinobacteria, proteobacteria etc. the microbial
capacity inside the gastro-enteric tract is about 10₁₂ CFU/ml to 10⁷
CFU/ml in the intestine and 10₁₄ CFU/ml in the colon. Due to the
lesser oxygen consumption in the upper intestine itself present gram
(+) ve coccus such as Streptococcus sp. but the anaerobic bacteria in
the intestine and colon are like Clostridium sp. Luminal epithelium
and mucus epithelium consist of microorganisms based on their
mucus degradation. Bacteroidesfragilis, Bifidobacteriumbifidiumare
the most specific bacteria within mucus layer and it utilizes glycans
body as a source of glycosidase, sulphatase and sialidase enzymes.
Beyond the colony formation of commensal bacteria in the gastrointestinal
tract by glycan formation further targeted on the less
polysaccharide content and the necessary host pathogenic mucin as
an energy resource in the gut microbiome. [8]
Review of Literature
Gut Microbiota in relation to Unconventional T Cell Ligands and Immune Modulation:
Multiple types of PRRs and TLRs, Nodded receptor, C- type lectin
receptors helped to suppress diverse foreign molecules in diversified
spatial or dynamic spectrum along with polar virus containing
nucleic acids to adhering bacterial lipoproteins as in extracellular
or intracellular environment. With similar structural and functional
properties host pathogenic interaction proceeds to rapid activity of
innate leukocytes. PRR ligands are specifically non self, ubiquitous
structurally similar as microbial contents such as LPSs, TLR4 ligand or
peptidoglycan agonist and virus or bacteria stimulatory nucleic acid
ligands. Individual PRR ligands can possess as stringent structural
properties to suppress wide variety of foreign molecules. However,
a large variety of cells can treat against the common microbial
pathogens that is an immediate response against host pathogenic
interaction. Biochemical pathways developed pathogenic interaction
to emphasize structural PRR ligands to recognize their necessary
functions. Efficient immune response can process host pathogenic
interactions with genetic diversity. [9]As an effective complement activation of PRR type recognition and
mammalian host derived foreign ligand recognition are contrary to
each other. MHC class I and II components proceed to peptidoglycan
and structural invariability. Structurally variable foreign molecules
are active in mutation as T cell receptor recognizes mutated peptides.
The unrequired and detrimental immune response activated by selfantigen
capacity by eliminating peptide responding naive T cells is
a negative selection. This Ag recognition by maintaining activated
lymphocytes has been recognized with limited protection with
known resource. Adaptive immunity and innate immunity are the
first line defense mechanism which critically specifies host pathogenic
interaction. [10]
Microbial regulation in the Gut responsive T cells through Interleukin- 6 produced by Enteric neurons:
Enteric neurons to prevent inducing iTreg:Crosstalk between
ENS neurons and Treg cells it is using ‘‘iTreg’’ system to induce
FoxP3+. Muscle layers surrounding myenteric plexus were detached
and inoculated into progenitor cell culture medium to activate ENS
in vitro. Inhibition of iTreg by enteric neurons caused sensitivity by
approximately 1/100 neurons per cultured CD4+ T cells. [1]Inhibition of iTreg by Enteric neurons through cytokine
factors:Expression of T cell mediated receptors inhibit iTreg to
distinguish neurons and T cells. Cytokine receptors added to iTreg
inducing cultures to induce T cell death through diethylenetriamine.
But in classical pathway neurotransmitters or neuropeptides were
not inducing the inhibition. Through gene expression the neurons
distinguish between iTreg cells from Foxp3 reporter mice with or
except neuron assimilated co cultures. [12]
Microbial assimilation on the neuron-Treg axis: Commensal
gut microbes such as Clostridium ramosum induces RORγ+ Treg
cells thus triggers neurons in vitro. The effect of mono colonization of
GF mice on ENS structure and composition while high Treg inducer
C. ramosum or non-inducer Peptostreptococcusmagnus affecting
immunofluorescent imaging by colon segments like as antibodies
recognized neuron cell bodies. [13]
Commensal bacteria by accumulating peripheral regulatory T cells produces metabolites:
ExtrathymicTreg cells were generated through microbial
metabolites in the specific pathogen free (SPF) mice but not
antibiotics treated microbiota deficient mice or germ free (GF)
treatment of mice. Few contaminated Treg cells expanded in CD4+
T cell population it is more effective in less Foxp3 containing mice.
ExtrathymicTreg cell was involved in vivo by promotion of butyrate
in antibiotic treated mice or in untreated SPF mice. Treg cell was
increased in colonic lamina propia in CNS1 sufficient mice but not
in CNS1 deficient mice thus it suggests that bacterial metabolite also
suppress prominent Treg cell populations through Foxp3 protein
stabilization. Treg cell population in butyrate treated mice is greater
in Foxp3 protein concentration than butyrate free cultures. The ability
of T cells and DC cells in preparing Treg cells in vitro the butyrate
expression levels were non-mutually exclusive. Direct Treg cell
promotes on CD4+ T cells to prosecute DCs by butyrate facilitation
and differentiation. Treatment with DCs was able to facilitate Foxp3
expression in naïve T cells precursored by CD3 antibody and TGF- β
except butyrate expression. Upon testing HDAC inhibitory activity
two different types of HDAC inhibitors with distinct chemical nature
a butyrate derivative known as phenylbutyrate used as a standard
for controlling the experiment. However, through HDAC inhibition
TSA and butyrate assimilated Treg cell was proceed for further DCs
treated binding with butyrate and optimum TSA. [14]GPCR signaling mediated Cross Talk between metabolism and inflammatory response:
ExtrathymicTreg cells were generated through microbial
metabolites in the specific pathogen free (SPF) mice but not
antibiotics treated microbiota deficient mice or germ free (GF)
treatment of mice. Few contaminated Treg cells expanded in CD4+
T cell population it is more effective in less Foxp3 containing mice.
ExtrathymicTreg cell was involved in vivo by promotion of butyrate
in antibiotic treated mice or in untreated SPF mice. Treg cell was
increased in colonic lamina propia in CNS1 sufficient mice but not
in CNS1 deficient mice thus it suggests that bacterial metabolite also
suppress prominent Treg cell populations through Foxp3 protein
stabilization. Treg cell population in butyrate treated mice is greater
in Foxp3 protein concentration than butyrate free cultures. The ability
of T cells and DC cells in preparing Treg cells in vitro the butyrate
expression levels were non-mutually exclusive. Direct Treg cell
promotes on CD4+ T cells to prosecute DCs by butyrate facilitation
and differentiation. Treatment with DCs was able to facilitate Foxp3
expression in naïve T cells precursored by CD3 antibody and TGF- β
except butyrate expression. Upon testing HDAC inhibitory activity
two different types of HDAC inhibitors with distinct chemical nature
a butyrate derivative known as phenylbutyrate used as a standard
for controlling the experiment. However, through HDAC inhibition
TSA and butyrate assimilated Treg cell was proceed for further DCs
treated binding with butyrate and optimum TSA. [14]GPCR signaling:G protein coupled receptor (GPCRs) are
an extensive family of membrane receptor factors a part seven
transmembrane α- helix domains with intracellular C terminal and
extracellular N terminal. Canonical forms of GPCRs are containing
intracellular region ligands, G proteins that targeted downstream
signaling and β- arrestins that controls excessive activation.
Occasionally NLRP3 can be interacted via GPCRs through different
metabolites, ions, neurotransmitters, hormones such interaction
triggers cross talk between metabolism, GPCR and NLRP3 signaling
and inflammatory activities. Though acetate leads to GPR43
dependence on inflammatory activities through K+ efflux, Ca2+ influx
and downstream hyperpolarization. In contrary acetate correlates
with GPR43 in bone marrow derived macrophages (BMDMs) which
suppress NLRP3 inflammasome via Ca2+ machinery through seizing
of Ca2+ mobilization. [15]
Host-Microbiota Communication:Gut microbiota has
commensal interaction with the microbes and enteric environment
and metabolic activities in the gut are sensibly changing in the
microbiota and gave signal transduction for regulating cellular
mechanism including host PRRs such as inflammatory activation.
In microbiota regulated metabolism inflammasomes were subjected
upon regulation by metabolites NLRP6, NLRP3 and pyrin. In the
higher expression of NLRP6 in the intestine ligand activities with
bacterial components have been shown in the gut. Metabolomics and
metagenomics studies in the caecum subjected to microbiota related
metabolism and NLRP6 inflammatory activation with downstream
epithelial IL-18 metabolism and development of microbiota
composition NLRP6 inflammatory protection. [16]
The complement system:The complement anaphylatoxin
receptor in myeloid cells processed into NLRP3 inflammatory
activation. ATP inflammasome complement axis is corresponded
with DAMPs by inflammatory activation to promote sterile
inflammation. High concentration of metabolite controls cholesterol
metabolism and homeostasis by precipitating cholesterol crystals
for inflammasome and pathogenicity in atherosclerosis. Autocrine
complement activity initiates metabolic reprograming to activate
T cell regulation and inflammation. Functional complement
metabolism inflammasome axis specifically indicates T cells. [17]
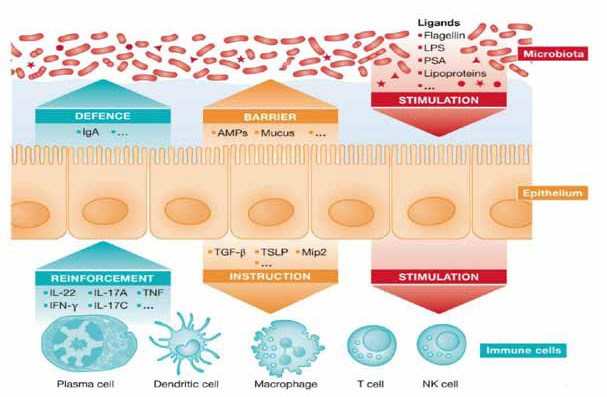
Innate immune signaling in intestine for epithelial homeostasis and disease:
Cell polarization and receptor signaling:Intestinal epithelial
cells have the polarity to express cell to cell contact to distinguish
plasma membrane into the apex and the basal region to facilitate
exo and endocytic metabolic trafficking. Enteric microbiota and
environmental nutrient stimuli protect the gut against microbial
attack. The stimulus expression is highly suppressed in basal bodies
of polarized human epithelial cell and murine mucosa cells in colon
by exposing into luminal flagellin on disrupted epithelial barrier. [18]Homeostasis and barrier integrity:Less innate immune signal
transduction with lack of epithelial specificity in signal molecules and
an expressed (-) ve form of MyD88 generates a barrier dysfunction
along with mucosal damage and inflammasome. Spontaneous
inflammation and Tnfdependant epithelial apoptosis were identified
in epithelial specific Tak 1 lesser mice. In contrary to the MyD88
initiated proinflammatory signaling the sub-epithelial immunity on
non haematopoetic cell signaling protects from homeostasis and
inflammation. [19]
Epithelial barrier integrity on mucus epithelium:Most frequent
model of mucus epithelium stabilizes and facilitates in penetration of
commensal bacteria into the gut. Epithelial specific deletion and NLRs
correspondingly support mucus level host defense. DSS induction on
inflammatory aggravation and epithelial deletion of NF- ҡβ subunits
thus downstream dominant signaling pathways have been activated.
[20]
Innate immunity in the Human Epithelium:Intestinal epithelial
gene expression helps in innate immunity in humans. Individuals with
lack of genetic compatibility suffers from diarrhea, colitis that was
not triggered by bone marrow transplantation. Homozygote human
allele have abnormal Paneth cells. Human polymorphism expression
is correlated with inflammatory bowel disease and also dislocation
of epithelial barrier is directed to detailed cell specific analysis. [21]
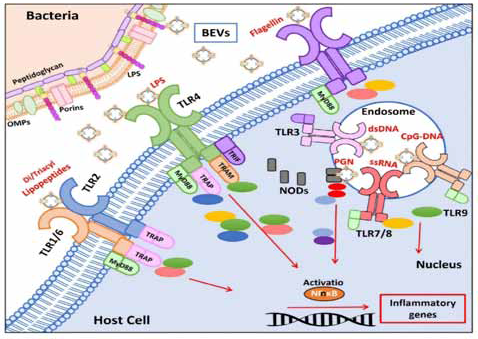
Bacterial membrane vesicle which is extracellular in nature:
Extracellular vesicles in Vibrio choleraehave been shown under
electron microscopy which was growing in log phase of bacterial cells
and from outer membrane it secrets spherical membrane structure
bound with the membranous periphery. Toxin secretion system
and vesicle type particles are considered as a structural articulation.Bacterial cell physiology is related with the structural membranous
vesicle and their biogenesis, compositions and functional efficacy.
Bacterial membrane vesicles are consisting of lipid bilayer which is a
nano particle in nature. Mainly gram (+) ve and gram (-) vebacteria
are composed of lipopolysachharides (LPS), peptidoglycans, protein,
lipid, nucleic acid etc. Direct contact of intracellular compound
in delivery of bacterial composites for the activation of bacterial
metabolites are necessary for membranous vesicle extra and
intracellular activities. [22]
Bacterial extracellular vesicles are associated with the
Biogenesis of membranous components:Extracellular cell wall
turnover rate are the leading cause of conditional production
of bacterial extracellular vesicle. Peptidoglycan fragments or
misfolded proteins causes the periplasmic outer membrane to
protrusion. Antibiotic ciprofloxacin assisted with the high potency
in heterogenous population of bacterial extracellular vesicles.
Cationic concentration and electronegative LPS resulted into local
negative charges which is further proceeds to coupling and repulsion
between LPS molecule and thus distortion of bacterial membrane and
vesicular detachment happened. Previous research articles suggested
that membrane phospholipid has been transferred between the outer
and inner vesicles that in turn causes membrane shrinkage and
lipopolysachharides have been triggered into outer vesicle membrane
through an acetylated disachharide named as lipid A. the gram (+)
ve bacterial extracellular vesicles transferred through porous cell wall
through cell membrane generated turgor pressure. P. aeruginosa cells
swelled up and burst and Bacillus subtilis released CMVs through
porous cell wall. Staphylococcus aureus bacterial cell wall releases
phenol soluble modulins and autolysins proteins which increases
membranous fluidity thus proceeds to production of CMVs. [23]
Formulation of gut immunity and activation of immune
response:The commensal bacteria and probiotic regulate host
immune response on bacterial extracellular vesicle to activate immune
cells through intestinal epithelium for interacting microbiota derived
immune cells on bacterial extracellular vesicles. [24]
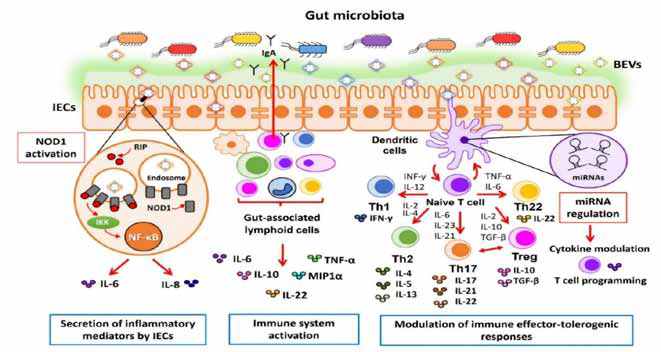
Microbiota derived immune response on bacterial extracellular
vesicle through intestinal epithelium: Commensal bacteria has been
imposed for the in vivo controlling of immune system in the intestine.
The cross talk between the microbiota, intestinal epithelium and the
immune system is considered as a human monolayer IECs stimulation
in the epithelial barrier and leads to the growth of immature dendritic
cells in the baso-lateral chamber to directly involve with the immune
system of lamina propia. [25]
Activating of immune cells through microbiota derived
Extra Cellular Vesicles:To balancing the immune system in gut
endothelium it must stimulates extracellular antigens and target
antibodies to provide barrier against host pathogens and infections.
i) Bacteroides sp.: The commensal bacteria B. fragilis benefitted
on the bacterial extracellular vesicle in food metabolism and
the gut ecosystem. Capsular polysachharide containing B.
fragilis is administrated against colitis and suppress immunity
of pro inflammatory and anti-inflammatory cytokines.
ii) Escherichia coli :Probiotic E. coli provides balance between
Figure 3:Bacterial Extracellular Vesicles associated molecular pattern recognition by host cell immune receptors
intestinal homeostasis and microbiota environment. The
immunomodulatory effects are associated with gut microbes
E. coli on immune activity of DCs and other innate immune
response. The differential expression of the miRNAs is
assimilated by triggering bacterial extra cellular vesicles on
DCs.
iii) Gram (+) ve commensal bacteria Lactobacillus sp.
and Bifidobacterium sp:Due to health benefits and
immunomodulatory effects bacterial extracellular vesicles
of Lactobacillus rhamnosus has been well studied for
probiotic functionality and neuron stimulating activities
by activating host pathogenic nervous system in enteric
epithelial system. Similarly,Bifidobacteriumbifidum targeted
a tolerogenic response by subcellular functions of probiotic
on the dendritic cells to prove immunomodulation mediated
bacterial extracellular vesicles. [27]
Role of probiotic immune system on gut microbiota
Helicobacter pylori:Intestinal immune system varies between
distinct mutualistic and symbiotic microorganisms from host
pathogenic activities to the tolerance level of commensal bacteria.
Gastrointestinal microbiota may cause to persistent and metabolic
disorders. In contrary to Helicobacter pyloriintestinal epithelium can
produce a physiochemical barrier to the prevention of pathogenic
colonization on the surface of mucus epithelium to recreate immune
tolerogenic factors against commensal bacteria. [28]
Microbial Dysbiosis:Commensal bacteria are the key providers
of energy resources for the host cellular enterocytes, inhibiting
pathogen colony formation, protecting against lymphoid tissue and
directing the immune response. Modulating the host pathogenic
response, formulating cell signaling, diminishing epithelial cell
polarity, altering gastric ulcer are the key primary resources on
alternation of gut microbiota in H. pylori infection. [29]
H. pylori and gut epithelium:Though H. pylori insisted
gastric microbiota has been preferred for strain specificity and host
pathogenic colony formation. The influence of H. pylori on gastric
microbiota has been suggested as Bacteroidetes regulated elimination
of H. pylori infection. This pathogen initiates activity of nuclear
factor kappa B (NF- ҡβ) proceeds to transcription factor stimulated
activities of MCP-1 from epithelium to monocytic extraction and
monocytic activation by LPS correlation with TL4. H. pylori infection
initiates excessive secretion of pro inflammatory cytokines such as
iNOS, TNF- α, IFN- γ , IL-8, IL-6, IL-4, IL-1β. [30]
Conclusion
Beyond relevant research publications on gut microbiota intrinsic
and extrinsic functionality of microbiome system and stimulation
of receptors, regulation of effectors to the target cells yet has to be
explored. These research findings are very much important for
developing translational mechanisms of microbiota derived diseases
in human health. The potency of applications of microbiota and
bacterial extracellular vesicles is very essential as a treatment purpose
or to overcome therapeutic challenges upon intestinal infections,
immune disorders and inflammation. The compositional diversity
of gut microbiome in a great extent to disease including oncogenic,
neurological, metabolic and immunogenic diseases has been essential
for gut microbiome profiling as a diagnostic tool.
Current researchers have been suggested that the next generation
sequencing in probiotics is intentionally targeted and genetically
modified organisms has been promoted to the beneficial recombinant
DNA technology for probiotic supplementation in gut microbiome
for its pharmaceutical output. However,another strategy is the
implementation of nanotechnology with micro-capsulation end point
it may develop the probiotic integrity on human health system thus
provide a thematic framework to eliminate the metabolic hazards in
probiotic supplementation.
Acknowledgments
The authors are thankful to Chancellor, Techno India University,
West Bengal, India for providing the necessary infrastructural
support for carrying out the research work.