Research Article
Evidences Indicating the Involvement of Selection Mechanisms for the Occurrence of C34 Anticodon in Bacteria
Vinod Kumar Prajapati1#, Siddhartha Sankar Satapathy2, Mattaparthi Venkata Satish Kumar1, Alak KumarBuragohain1,3 and Suvendra Kumar Ray1*
Corresponding author: Dr. Suvendra Kumar Ray, Department of Molecular Biology and Biotechnology, Tezpur University,Napaam, Tezpur-784028, Assam, India, Tel: (+91) 3712 275406; Fax: (+91) 3712 - 267005/267006; E-mail: suven@tezu.ernet.in
Citation: Prajapati VK, Satapathy SS, Satish Kumar MV, Buragohain AK, Ray SK. Evidences Indicating the Involvement of Selection Mechanisms for the Occurrence of C34 Anticodon in Bacteria. J Cell Sci Molecul Biol. 2015;2(1): 112.
Copyright © 2015 Suvendra Kumar Ray et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Submission: 23/03/2015; Accepted: 07/04/2015; Published: 12/04/2015
Abstract
To decode all the 61 sense codons in the genetic code, the number of different anticodons used in bacteria varies between 24 to 45. If a small set of anticodon can decode all the codons in a bacterium, the evolutionary significance of the increase in anticodon diversity in a bacterium is not understood well. The higher anticodon diversity in G+C% high genomes has been attributed mainly to the occurrence of many C34 anticodons (C at the 1st anticodon position) in bacteria. Whether C34 anticodons are restricted only to the bacteria with high genome G+C% or occurrence of some of these is also extended to some bacteria with low genome G+C% has been addressed in this study. We have analyzed the occurrence of C34 anticodons in 201 bacteria that represent all the major taxa and also covered a range of G+C% from 20 to 73. Our analysis suggests preferential occurrence of C34 anticodons with respect to the codons of some amino acids (Leu, Arg) such as UUR, AGR even in bacteria with low G+C% genomes. On the other hand such preferential occurrence of C34 anticodon was not seen in the cases of the codons for some other amino acids (Ala, Val) such as GCN, GUN even in bacteria with high G+C% genomes. The findings in this study indicate that the occurrence of C34 anticodon is influenced by some selection mechanism in bacteria. Based on the findings we have discussed the role of near-cognate tRNA during translation and a possible role of translation kinetics influencing the occurrence of C34 anticodons in bacteria.
Keywords: Transfer RNA, C34 anticodons, Anticodon diversity, Sparing strategy, Genome G+C%, Codon-anticodon interaction
Introduction
During translation, due to wobble base pairing that occurs between the 3rd codon position and the 1st anticodon position (34th nucleotide in tRNA), one tRNA can decode more than one codon and one codon can be decoded by more than one tRNA [1]. Therefore, lesser number of anticodons is used to decode the 61 sense codons of the genetic code. On the basis of the known wobble base pairing rule, 24 anticodons are sufficient for carrying out translation unambiguously. However, it has been reported that the number of anticodon deployed in bacteria may be as high as 45 [2]. The nearly two fold variation in the occurrence of different anticodons in bacteria, termed as anticodon diversity, is attributed to the presence of G34 anticodon for the eight family box codons and C34 anticodons for the thirteen NNR codons which includes the eight family box codons in the genetic code. Deployment of higher number of anticodons for execution of the translation process is not clearly understood in the context of the theoretical requirement of 24 anticodons for the purpose. Though an unmodified U34 anticodon can decode all the four synonymous codons in a family box, often modified U34 anticodon is observed in bacteria that restricts its decoding ability to three out of the four synonymous codons in a family box [3]. In this case the occurrence of G34 for the eight family box codons is not redundant in many bacteria as G34 anticodon would be essential for decoding the C-ending codon of the family box. The selection force behind the evolution of U34 anticodon modification of family box tRNA is yet to be understood completely though it has been reported that the modification of U34 is important for translational selection of codon usage bias in bacteria [1]. Unlike the G34 anticodon, the occurrence of the thirteen C34 anticodons is apparently redundant in bacteria. This is because C34 anticodons decode G3 codons in a bacterium, which already possesses U34 anticodons that decode the G3 codons as well. Therefore understanding the occurrence of C34 anticodon in bacteria is of great significance from the evolutionary point of view.
Earlier it has been reported that anticodon diversity positively correlates with the genome G+C% in bacteria [4,5]. The correlation result is well explained on the basis of anticodon occurrence to decode family box codons in bacteria [6]. For example, in bacteria with very low genome G+C%, only the U34 anticodon (anticodon with U at the 1st position) is used for decoding all the four synonymous codons in a family box. This is referred to as sparing strategy #3 [6]. In bacteria with high genome G+C%, U34, G34 and C34 anticodons are used to decode the same four synonymous codons. This is referred to as sparing strategy #1. In some other bacteria, U34, and G34 anticodons are used for decoding the four synonymous codons in a family box, which is known as sparing strategy #2. Generally the use of sparing strategies and genome G+C% are inversely related in bacteria. In case of Lys, Gln, Glu, etc, that are encoded only by the NNR codons, generally U34 and C34 anticodons occur in the bacteria with high G+C% genomes while in the bacteria with low G+C% genomes only U34 anticodon occurs. The advantage of C34 anticodons to the bacteria with high G+C% genomes is not fully understood.
It is known that genome G+C% and codon G+C3% (codon 3rdposition G+C%) positively correlates [7,8,9]. There is no report on such correlation between G+C% at different anticodon positions and genome G+C%. Since an anticodon comprises of only three nucleotides in a tRNA, its occurrence might be thought of less significant in the context of genome G+C% unlike codons that occur many times in a genome. However, a single tRNA species is present many times in a cell as a transcript. Therefore, anticodon G+C% is also likely to be influenced by genome G+C% like codon G+C%. So the increase in C34 anticodons in G+C% high genomes can be argued in favor of G+C%. It is pertinent to note that the influence of genome composition on N36 position in anticodons (N at the 3rd anticodon position) can also be observed in bacteria [3]. Arg and Leu are encoded by both G+C% low codons (AGRArg, UURLeu) and G+C% high codons (CGNArg, CUNLeu). Decoding of these G+C% low codons is done by either the A36 or the T36 anticodons whereas decoding of the G+C% high codons is done by the G36 anticodon [3].
Though anticodon diversity influences translation, a fundamental process in cell, only a few reports have addressed this evolutionary question in bacteria [10,3,1,5]. To find out the significance of the occurrence of the anticodon in bacteria, In this study we compared C34 occurrence across genomes of different G+C%.
Results
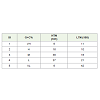
Anticodons of 201 bacteria were studied from the tRNA genomic database. (http://gtrnadb.ucsc.edu/) [11]. The same list of bacteria was used earlier by Wang et al [3] for a different purpose. The list included bacteria belonging to all the major Classes and subclasses (Supplementary Table 1). The bacteria were further divided into HTN group (those which had total tRNA gene number more than equal to 50) and LTN group (those which had total tRNA gene number less than 50) (Supplementary Table 1). In HTN total 101 bacteria and in LTN total 100 bacteria were included (Table 1).
Correlation study was made between the anticodon G+C% and the genome G+C%. The correlation was positive (Pearson r value 0.81) between the genome G+C% and the anticodon G+C%. Correlation between the genome G+C% and the anticodon G+C% at the 1st (G+C1%), the 2nd (G+C2%), and the 3rd (G+C3%) positions were also positive (Pearson r value 0.78, 0.38, 0.66, respectively) (Table 2A). To understand the contribution of the anticodon diversity towards the above correlation result, the correlation study was carried out without including the anticodons for Met, Trp, Phe, Tyr, Cys, His, Asn, Asp where anticodon diversity did not occur. Anticodon of Ile, where anticodon diversity is generally not variable along genome G+C%, was also not included. The correlation was also strong (Pearson r value 0.82) even without including the anticodons of the above amino acids (Table 2). Correlation between the genome G+C% and the anticodon G+C% at different positions was also positive (Pearson r value 0.78, 0.26, 0.57 respectively) (Table 2). It is known in bacteria with high G+C% genomes that the codons with high G+C%, such as those for Arg (CGNArg) and Leu (CUNLeu) are used more frequently than the colons with low G+C% like those for Arg (AGRArg) and Leu (UURLeu). The reverse is also true in bacteria with low G+C% genomes (Palidwor et al 2012). Therefore, further correlation study was carried out between the anticodon G+C% and the genome G+C% without including the anticodons for Arg and Leu (Table 2). The correlation between anticodon G+C% and genome G+C% was negative (Pearson r value -0.65) unlike the above two correlations. But the correlation between anticodon G+C1% and genome G+C% was positive (Pearson r value 0.79) and the negative correlation was due to negative correlations between G+C2% and genome G+C% (r value – 0.67) as well as G+C3% and genome G+C% (r value – 0.66) (Table 2). In ummary, the correlation studies between the anticodon G+C1% and the genome G+C% in the bacteria considered in the present study ndicated that there was an increase in the G34 and the C34 along with the genome G+C%.
The bacteria were divided into five different groups on the basis of their genome G+C%. As the maximum G+C% known in bacterial genomes was 75%, five different groups were made ranging from the very high G+C% to the very low G+C% genomes as follows. The very high G+C% (VH) group included bacteria with genome G+C% ≥65.00; the high G+C% (H) group included those with 65.00> G+C% ≥55.00; the moderate G+C% (M) group included bacteria with 55.00> G+C% ≥45.00; the low G+C% (L) group with 45.00> G+C% ≥35.00; and the very low G+C% (VL) group included bacteria with 35.00> G+C% (Table 1). This grouping was done for a comparative analysis of the C34 occurrence in different G+C% groups.
The occurrence of each of the thirteen C34 anticodons, which included the eight family boxes and the five NNR codons (Table 3 ) in various G+C% groups both under the HTN and in the LTN, was studied. In the different G+C% groups, the % of bacteria having a particular C34 was analyzed (Table 3). As expected from the correlation result, C34 occurrence was most frequent in the VH group and gradually decreased towards the VL group in each of the thirteen cases with a few exceptions. However, it was interesting to observe the large variation in C34 occurrence in the different amino acids within a particular G+C% group. For example in the M group of the HTN which comprised of 36 different bacteria, C34 anticodon of Leu (LeuCAA) was occurring in all the bacteria (frequency value was 1.0) whereas C34 anticodons of Ala (AlaCGC) and Val (ValCAC) Leu (LeuCAA) were found to have few occurrence (frequency was 0.0 and 0.08, respectively). In the L group of the HTN comprising of 37 different bacteria, C34 anticodon of Leu (LeuCAA) were found to occur again in all the bacteria (frequency value was 1.0) whereas C34 anticodons of Ala (AlaCGC) and Val (ValCAC) Leu (LeuCAA) were found only in a few bacteria (frequency was 0.0 and 0.05, respectively).The observation indicated a general avoidance of C34 anticodons for Ala (AlaCGC) and Val (ValCAC) in contrast to a preferential trend to possess C34 anticodons for Leu (LeuCAA). In fact, occurrence of LeuCAA was found in most of the bacteria in the different G+C% groups in the HTN and in the LTN. Differential occurrence was also observed with respect GlyCCC, ProCGG, ThrCGU and SerCGA within a G+C% group. Similarly, there were differences among GlnCUG, GluCUC and LysCUU within a G+C% with respect to their occurrence. This observation of C34 occurrence was similar for the HTN and the LTN. The amino acid specific variation in C34 occurrence was evident in the heat-map dendrograms (Figure 1). The occurrence of C34 in the VL group for some amino acids and the avoidance of C34 in the VH group for some amino acids indicated the influence of some selectionmechanism.
Figure 1a: a(i) and a(ii): Heat map dendrogram of C34 anticodon occurrence in bacteria with high tRNA number and low tRNA number. There ae total 13 cases where NNR codons coding for the same amino acids and for each case C34 occurrence is possible. Amino acids have been represented by their single letter code in the X-axis. Leu has been grouped into L_4 and L_2, similarly Arg has been grouped into R_4 and R_2. From the heat map it is evident that C34 anticodons are more preferred for some amino acid codons such as and for some other amino acid codons C34 anticodons are less preferred in bacteria. As described in the text, Ala and Val where C34 occurrence chance is lower have been grouped together. Similarly, Arg and Leu where C34 occurrence chance is higher, have been grouped together. The grouping of amino acids in HTN (i) and LTN (ii) are not same. But the conclusion from the study that C34 occurrence is found in low G+C% genomes is true for both HTN and LTN.
Figure 1B: b (i) and b (ii): Heat map dendrogram of G34 anticodon occurrence in bacteria with high tRNA number and low tRNA number. There ae total 08 family boxes in the genetic code where G34 avoidance is possible. Amino acids have been represented by their single letter code in the X-axis. From the heat map it is evident that G34 anticodon is most preferred for Gly and while for Arg G34 anticodon is least preferred in bacteria. The grouping of amino acids in HTN (i) and LTN (ii) are not same.
The occurrence of G34 across the eight family box anticodons in different G+C% groups in the HTN and in the LTN was analyzed (Table 3). G34 occurrence was found to be more in the high G+C% bacteria and less in the bacteria with low G+C%. However, the extent of difference between the high G+C% and the low G+C% bacteria with respect to G34 occurrence was lower than that with respect to the occurrence of C34. This indicated that G34 occurrence was more essential than C34 occurrence in bacteria. The general bservation was that C34 occurrence was often accompanied with G34 occurrence but the reverse was not true. G34 occurrence for GlyGCC was observed to be prevalent in all the G+C% groups while in case of AlaGGC and ProGGG, its occurrence was lower in the G+C% low group. Among the eight amino acids within a G+C% group there was difference in the occurrence frequency of G34. The amino acid specific variation in G34 occurrence was evident in the heat-map dendrograms (Figure 1). The low frequency of G34 in AlaGGC and ProGGG and the high frequency of G34 in GlyGCC, and ThrGGU within a G+C% group indicated the influence of a selection mechanism for its occurrence Table 4.
In case of the Arg family box, neither C34 nor G34 occurrence was discussed because unlike the other family boxes, C34 occurrence is essential here to decode the CGG codon as U34 anticodon for this family box is usually absent in bacteria. In bacteria the anticodon modifying enzyme for tRNAArg family box is different in the sense that A34 (adenine) is converted to I34 (inosine) [6].
A study was made to compare the copy number of U34, G34 and C34 anticodon genes among different amino acids. The occurrence of U34 gene numbers across different G+C% was found to be similar indicating low influence of the genome G+C% on its occurrence. As expected C34 and G34 gene numbers were more in the high genome G+C% bacteria. But C34 and G34 gene numbers were either one or two and lesser than that of U34 even in the G+C% high genomes. The exception was G34 tRNA gene numbers of Gly where the gene number was 3 to 4 even in the low G+C% genomes. In fact G34 gene number was more than U34 gene number in case of Gly. Box plot of the C34 and G34 gene numbers for different amino acids also suggested the difference among different amino acids (Figure 2). In case of Ala, Val and Glu, it was evident that C34 gene number was low. In general lower gene number of C34 anticodons than U34 and G34 anticodongene numbers in the high G+C% bacteria indicated weak selection on this anticodon in G+C% high bacteria.
Figure 2: A box plot diagram for the tRNA gene numbers having C34 anticodon (Figure 2a) for the tRNA gene numbers having G34 (Figure 2b)anticodon in the HTN groups. There are total 13 cases where C34 gene number have been (Fig. 2a) considered. Three letter code of amino acids have been written. It is very evident that Leu4, Leu2, Arg4, Arg2 cases have higher gene number of tRNA with C34 anticodons, whereas lower gene numbers of tRNA with C34 anticodons for Val, Ala and Glu. This is in concordance with the finding from the heatmap result. Glu_C34, Ala_C34 and Val_C34 sample data are less significant than the rest of the samples (Supplementary Table 2). There are total 08 cases where G34 gene number have been (Figure 2b) considered. Three letter code of amino acids have been written. It is very evident that Gly has the higher gene number of tRNA with G34 anticodons.
Discussion
In this study we addressed the issue of the importance of increased anticodon diversity in bacteria. Anticodons are so fundamental totranslation, its diversity among bacteria is a very exciting which may shed light on the mechanism of molecular evolution. Considering the wobble pairing rule, C34 occurrence is apparently redundant in the presence of U34 in a genome as the latter can wobble pair with G3. Occurrence of C34 is more prevalent in the high G+C% genomes than in the low G+C% genomes. This is in concordance with the view of neutral theory of evolution which suggests that the occurrence C34 in genomes is a mere function of the genome composition which favors reater increase in anticodon G+C%. So, low occurrence of C34 in some bacteria need not affect translation. From selectionist point of view, it is belived that C34 occurrence is required in bacteria as U34:G3pairing is not strong during translation [3]. As the G3 abundance is more in the high G+C% genomes than in the low G+C% genomes, C34 occurrence is prefereed in the former. Based on this argument which favors selection theory of evolution, C34 occurrence is expected to occur more frequently in the high G+C% genomes. If there is indeed translational selection for C34 anticodon in G+C% high genomes, then it should be more prominent for amino acids where G3 codons are under more selection than the other synonymous codons in thehigh expression genes. Considering the argument of the translational selection, anticodon diversity might also be observed in some G+C% low genomes with translational selection is high on G3 codons in the high expression genes.
Though in general C34 occurrence was observed more in the high G+C% bacteria (Satapathy et al [4]; and also this study), this study revealed certain exceptions relating to C34 occurrence that was reported earlier: (i). the C34 anticodon in case of Leu (LeuCAA) was observed even in the very low G+C% genomes ; (ii). the absence of C34 anticodon in case of Ala (AlaCGC) and Val (ValCAC) even in some bacteria with high G+C% genomes. Our study indicates that that there are some codons against which C34 anticodon is preferred and there are some codons against which occurrence of C34 anticodon is not preferred. Therefore it may be inferred that genome composition and U34:G3 weak pairing are not sufficient reasons to explain the occurrence of C34 in bacteria. Below we are forwarding some additional possible explanations for the occurrence of C34 anticodon in bacteria.
It is likely that U34:G3 pairing is not always weak. Strength of the pairing at the wobble position might be influenced by the base pairing at codon posion 1 and 2. In case of Ala and Val, U34:G3 pairing is likely to be strong due to the influence of the base pairing at codon posion 1 and 2. This might be the reason for the avoidance of AlaCGC and ValCAC in the high G+C% bacteria. In cas of Leu, the LeuCAA anticodon decodes the UUG codon in bacteria which is more abudant in the low G+C% genomes than the other synonymous codons. The occurence of the LeuCAA anticodon in genomes might be argued in view of the inefficient decoding of the UUG codon by the anticodon LeuUAA. The presence of the UA dinucleotide as a part of the anticodon LeuUAA might be affecting the decoding process during translation as it has been reproted that UA is generally avoided in coding sequences in bacteria [13,14].
Apart from the efficient codon:anticodon interaction, the decoding kinetics of some codons during translation might influence the occurrence of C34 anticodon in bacteria [15]. Recent studies have provided evidence indicating that decoding rate of a codon is positively influenced by the cytosolic concentration of the cognate tRNA, and negatively influenced by the cytosolic concentration of near-cognate tRNA [16-19] Cognate tRNA is the one with correct anticodon constitution against a codon . Near cognate tRNA is the one with anticodon that can undergo Watson-Crick pairing with two nucleotides of a codon. For example, His-tRNA function as a near cognate tRNA of Gln and the vice versa is also true. In a more critical analysis isoacceptor tRNA can function as near cognate tRNA of a synonymous codon. Following is the example of Gly codon and Glyanicodon in this regard. To decode the four Gly codons, UCC and GCC anticodons are sufficient in bacteria. UCC anticodon decodes the GGG, GGA and the GGU codons while the GCC anticodon decodes the GGU and GGC codons. It is the cognate-tRNA : nearcognate- tRNA ratio that determines the decoding time of a codon during translation. Therefore, while the occurrence of the UCC anticodon delays the rate of GGC decoding, the occurence of the GCC anticodon delays the rate of the GGA and the GGG decoding during translation.Similarly while the CCC anticodon delays the decodingof the GGA, GGU and theGGC codons, its occurences expedites the decoding of GGG codon. So the occurrence of C34 anticodon delays the decoding of some codons while it also expedites decoding of some other codons during translation. The net advantage to the cell for these two opposite kinetics would be the determining factor for the C34 occurrence in bacteria . This might be the reason why we do not observe a significant increase in C34 anticodon gene numbers in the bacteria with high G+C%. It has been reported earlier that in the fast growing bacteria low tRNA diversity and high tRNA gene numbers are preferred (Rocha, 2004).
While U34 is sufficient to decode the family box codons, due to the requirement for more effcient decoding of codons in some of the family box codons, simultaneous evolution of U34 modifying enzyme along with G34 anticodon possibly had occurred. Both are required for efficient decoding of the U-ending codons in family boxes and the U-ending codons are observed to be selected in different family boxes in bacteria [20]. This might be the reason for the spraing strategy #2 in the bacteria with low G+C%. The observation of the higher G34 gene number in general for the Gly family box might be due to strong translational selection on the GGU codon [21]. In the high G+C% genomes, selection of the G-ending codon in family boxes is not usually observed though C34 anticodon occurs frequently in these genomes. This might be the reason why the C34 gene copy number is found to be lower than that of the U34 and G34 genes. So the occurrence of C34 in genomes might be the result of a combined effect of the genome G+C%, codon:anticodon pairing, expeditious decoding of some codons and delayed decoding of some other codons, with an overall advantage to the bacteria.
Our study has kept the question regarding evolution of C34 in bacteria open for speculation on molecular evolution of anticodon diversity. Future studies will reveal more interesting aspects on the evolution of anticodon diversity in bacteria.
Acknowledgements
VKP thanks DBT, Govt of India for the MSc fellowship and the grant for MSc thesis, Govt of India for the MSc fellowship and the grant for MSc thesis, and also for the Bioinformatics Infrastructure Facility (DBT-BIF) at Tezpur University.
References
- Ran W, Higgs PG (2010) The influence of anticodon-codon interactions and modified bases on codon usage bias in bacteria. Mol Biol Evol 27: 2129-2140.
- Satapathy SS, Powdel BR, Dutta M, Buragohain AK, Ray SK (2014a) Selection on GGU and CGU codons in the high expression genes in bacteria. J Mol Evol 78: 13-23.
- Osawa S, Jukes TH, Watanabe K, Muto A (1992) Recent evidence for evolution of the genetic code. Microbiol Rev 56: 229-264.
- Satapathy SS, Dutta M, Ray SK (2010a) Variable correlation of genome GC% with transfer RNA Number as well as with transfer RNA diversity among different bacterial groups: α-Proteobacteria and Tenericutes exhibit strong positive correlation in both cases. Microbiological Res 165: 232-242.
- Satapathy SS, Dutta M, Ray SK (2010b) Higher tRNA diversity in thermophilic bacteria: a possible adaptation to growth at high temperature. Microbiological Res 165: 609-616.
- Grosjean H, de Crecy-Lagard V, Marck C (2010) Deciphering synonymous codons in the three domains of life., co-evolution with specific tRNA modification enzymes. FEBS LETTs 584: 252-264.
- Muto A, Osawa S (1987) The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci USA 84: 166-169.
- Palidwor GA, Perkins TJ, Xia X (2010) A general model of codon bias due to GC mutational bias. PLoS ONE 5: e13431.
- Agashe D, Shankar N (2014) The evolution of bacterial DNA base composition. J Exp Zool B Mol Dev Evol 322: 517-528.
- Osawa S, Ohama T, Yamao F, Muto A, Jukes TH, et al. (1988) Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc Natl Acad Sci USA 85: 1124-1128.
- Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res 25: 955-964.
- Wang B, Shao Z-Q, Xu Y, Liu J, Liu Y, et al. (2011) Optimal codon identities in bacteria: implications from the conflicting results of two different methods. PLoS ONE 6: e22714.
- Karlin S, Campbell AM, Mrázek J (1998) Comparative DNA analysis across diverse genomes. Annu Rev Genet 32: 185-225.
- Satapathy SS, Powdel BR, Dutta M, Buragohain AK, Ray SK (2014b) Constraint on dinucleotides by codon usage bias in bacterial genomes. Gene 536: 18-28.
- Tarrant D, von der Haar T (2014) Synonymous codons, ribosome speed, and eukaryotic gene expression regulation. Cell Mol Life Sci 4195-4206.
- Rodnina M.V, Wintermeyer W (2009) Recent mechanistic insights into eukaryotic ribosomes. CurrOpin Cell Biol 21: 435-443.
- Kothe U, Rodnina MV (2007) Codon reading by tRNAAla with modified uridine in the wobble position. Mol Cell 25: 167-174.
- Gromadski KB, Rodnina MV (2004) Kinetic determinants of high-fidelity tRNA discrimination on The ribosome. Mol Cell 13: 191-200.
- Pape T, Wintermeyer W, Rodnina M (1999) Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J 18: 3800-3807.
- Rocha EP (2004) Codon usage bias from tRNA’s point of view, redundancy, specialization, and efficient decoding for translation optimization. Genome Res 14: 2279-2286.
- Satapathy SS, Powdel BR, Dutta M, Buragohain AK, Ray SK (2014b) Constraint on dinucleotides by codon usage bias in bacterial genomes. Gene 536: 18-28.